Single-molecule force spectroscopy on synthetic molecular rotors
M. Ledent1, D. Sluysmans1, D. Dattler2, Q. LI2, T. Ellis2, N. Giuseppone2 and A.-S. Duwez1
1 UR Molecular Systems, University of Liège.
2 SAMS research group, University of Strasbourg.
m.ledent@uliege.be
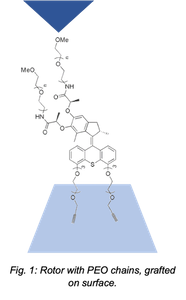
Here, we have investigated the mechanical response of these rotors by atomic force microscopy (AFM)-based single-molecule force spectroscopy. Rotors were grafted on a silicon surface through click chemistry and linked to the AFM tip by physisorption of two poly(ethylene oxide) (PEO) tethers (Fig. 1). The physisorbed PEO chains on the AFM tip allow us to probe singular behaviors of the rotor depending on their degree of entanglement. Pulling experiments on UV irradiated or unirradiated molecules evidenced different characteristic profiles under external force. The molecular rotation was investigated by comparing the rupture force values, the extensions at rupture and the single-molecule behavior of the polymers under load. Results were also compared to a reference molecule unable to rotate under UV light due to an episulfide bridged double-bond. Force clamp and pulling-relaxing experiments will provide new insights in the identification of other crucial single-molecule parameters such as the rotation speed, the generated work or the stall force.
1. For a review, see Cnossen, A., Browne, W. R. and Feringa, B. L. Top. Curr. Chem. 2014, 354, 139.
2. Li, Q., Fuks, G., Moulin, E., Maaloum, M., Rawiso, M., Kulic, I., Foy, J. T. and Giuseppone, N. Nature Nanotechnol 2015, 10, 161.
